An understanding of systems biology
provides an excellent paradigm for
the materials scientist. Ultimately one
would like to take an “atoms-applications”
approach to materials design.
This paper describes how the concepts
of genomics, proteomics, and other
biological behavior which form the
foundations of modern biology can be
applied to materials design through
materials informatics.
INTRODUCTION
…describe the overall significance
of this paper?
This paper describes how the
concepts of systems biology are
applicable to the field of materials
science and engineering.
…describe this work to a materials
science and engineering professional
with no experience in your
technical specialty?
This paper describes how
informatics is based on the use
of mathematics and modeling
strategies for linking length scales
in materials behavior.
…describe this work to a layperson?
The paper describes how the concepts
of genomics, proteomics, and other
biological behavior which form the
foundations of modern biology can
be applied to the design of materials
from the “atom to applications.”
|
The concept of complexity in biology
and how to assess the links between
information at the molecular level to
that at the living organism (e.g., genomics,
proteomics, etc.) is the foundation
of systems biology. The understanding
of systems biology provides
an excellent paradigm for the materials
scientist. Ultimately one would like to
take an “atoms-applications” approach
to materials design. How do we organize
atoms and build systematically
structural units at increasing length
scales to the final engineering component
or structure? At present we need
to rely on extensive prior knowledge
with experiments, computation, and
even failure analysis to understand the
complex network of interactions of materials
behavior which govern the performance
of an engineering system.
The problem is that even with advanced
experimental and computational tools,
the rate of discovery is still slow, only
punctuated by unexpected findings
(e.g., superconducting ceramics, conducting polymers) which stimulate new
areas of research and development. The
iterative approach as shown in this paper
is common to many fields as one
tries to link observations with models.
The challenge is to develop models that
capture the system behavior by accounting
for all the different levels of
information that contribute to the systems
behavior.
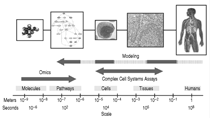
The goal of modern systems biology
is to understand physiology and disease
from the level of molecular pathways,
regulatory networks, cells, tissues, organs,
and ultimately the whole organism.2 As currently employed, the term
“systems biology” encompasses many
different approaches and models for
probing and understanding biological
complexity, and studies of many organisms
from bacteria to humans. A similar
paradigm exists for materials (e.g.,
atoms to airplanes).2,3
As aptly described by E.C. Butcher
et al.,2 the “-omics” (bottom-up) approach
focuses on the identifi cation
and global measurement of molecular
components. Modeling (the top-down
approach) attempts to form integrative
(across scales) models of human physiology
and disease, although with current
technologies, such modeling focuses
on relatively specific questions at
particular scales (e.g., at the pathway
or organ levels). An intermediate approach,
with the potential to bridge the
two, is to generate profiling data from
high-throughput experiments designed
to incorporate biological complexity at
multiple levels: multiple interacting active
pathways, multiple intercommunicating
cell types, and multiple environments.
A similar challenge occurs in materials
science, identifying pathways of
how chemistry, crystal structure, microstructure,
processing variables, and
component design and manufacturing
“communicate” with each other to define performance. This forms the materials
science equivalent of the biological
regulatory network.
COMPLEXITY IN MATERIALS SCIENCE AND BIOLOGY
Because biological complexity is an
exponential function of the number of
system components and the interactions
between them, and escalates at
each additional level of organization
(Figure 1), such efforts are currently
limited to simple organisms or to specific minimal pathways (and generally
in very specific cell and environmental
contexts) in higher organisms. The
same can be said of complexity in materials
science. Even if our ability to measure molecules and their functional
states and interactions were adequate
to the task, computational limitations
alone would prohibit the understanding
of cell and tissue behavior from the
molecular level. Thus, methodologies
that filter information for relevance,
such as biological context and experimental
knowledge of cellular and
higher level system responses, will be
critical for successful understanding of
different levels of organization in systems
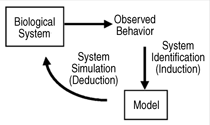
biology research. As described
by H. Kitano,5 a cycle of biological
research begins with the selection of
contradictory issues of biological signifi
cance and the creation of a model
representing the phenomenon. Models
can be created either automatically or
manually. The model represents a computable
set of assumptions and hypotheses
that need to be tested or supported
experimentally.
A similar analogy may be applied to
materials science in trying to explain
an unexpected or unusual materials behavior
such as the discovery nearly two
decades ago of high-temperature superconductivity
being exhibited by oxide
systems. Up to that point the majority
(but not all) of research in this field
was focused on intermetallics. The new
discovery at the time by Bednorz and
Mueller in 1986 spawned a vast array
of studies both experimental and theoretical
to gain a better understanding of
the causes of this important materials
behavior. This, of course, was part of a
cycle of hypothesis-driven research in
superconductivity that has had a long
and distinguished history.
The computational
simulations (biologists refer to
them as “dry” experiments) on models
reveal computational adequacy of the assumptions and hypotheses embedded
in each model. Inadequate models
would expose inconsistencies with established
experimental facts, and thus
need to be rejected or modified. Models
that pass this test become subjects
of a thorough system analysis where a
number of predictions may be made. A
set of predictions that can distinguish a
correct model among competing models
is selected for experimental validation
(called “wet” experiments by the
biologists). “Successful” experiments
are those that eliminate inadequate
models.
Models that survive this cycle are
deemed to be consistent with existing
experimental evidence. While this is an
idealized process of systems biology
research, the hope is that advancement
of research in computational science,
analytical methods, technologies for
measurements, and genomics/material
informatics can transform research to
fit this cycle for a more systematic and
hypothesis-driven science.
COMPUTATIONAL FOUNDATION FOR MATERIALS INFORMATICS
As suggested by T. Ideker and D.
Lauffenberger,6 relationships between
different components of information
can be extracted from the scaffold using
high-level computational models,
which identify the key components, interactions,
and infl uences required for
more detailed low-level models. Large-scale
experimental measurements validate
high-level models, whereas targeted
experimental manipulations and
measurements test low-level models.
The ultimate goal of knowledge discovery
is achieved in systematic integration
of data, correlation analysis developed
through data mining tools, and most
importantly, validated by fundamental
theory and experiment based science
of materials.
The sources of data can
be varied and numerous, including
computer simulations, high-throughput
experimentation via combinatorial
experiments and large-scale databases
of legacy information. The application
of advanced data mining tools permits
the processing of very large sets of information
in a robust yet rapid manner.
The collective integration of statistical
learning tools (the high-level models as
shown above) with experimental and
computational materials science allows
for an informatics driven strategy for
materials design.7–10
Ultimately the processing-structure-properties
paradigm which forms the
core of materials development is based
on understanding multivariate correlations
and their interpretation in terms
of the fundamental physics, chemistry,
and engineering of materials. The field
of materials informatics can advance
that paradigm in a significant manner.
A few critical questions may be helpful
to keep in mind in building the informatics
infrastructure for materials science.
How can data mining/machine learning
best be used to discover what attributes
(or combination of attributes) in
a material may govern specific properties?
Using information from different
databases, we can compare and search
for associations and patterns that can
lead to ways of relating information
among these different datasets.
What are the most interesting patterns
that can be extracted from the
existing material science data? Such a
pattern search process can potentially
yield associations between seemingly
disparate data sets as well as establish
possible correlations between parameters
that are not easily studied experimentally
in a coupled manner.
How can we use mined associations
from large volumes of data to guide
future experiments and simulations?
How does one select from a materials
library, and which compounds are
most likely to have the desired properties?
Data mining methods should
be incorporated as part of design and
testing methodologies to increase the
efficiency of the material application
process. For instance, a possible test
bed for materials discovery can involve
the use of massive databases on crystal
structure, electronic structure, and thermochemistry.
Each of these databases
by themselves can provide information
on over hundreds of binary, ternary,
and multicomponent systems. Coupled
to electronic structure and thermochemical
calculations one can enlarge
this library to permit a wide array of
simulations for thousands of combinations
of materials chemistries. Such a
massively parallel approach in generating
new virtual data would be daunting
if not impossible were it not for data
mining tools as proposed here.
CONCLUSIONS
The linking of length scales is a
pervasive theme in both biology and
materials science. Understanding the complexity across these length scales
requires a systems approach to both
disciplines. The gene-to-organ or atomto-
application paradigm for design can
serve as the “omics” approach for materials
science and informatics is the
enabling toolkit for this to occur.
ACKNOWLEDGEMENTS
K. Rajan acknowledges support
from the Office of Naval Research for
Multidisciplinary University Research
Initiative program: Novel Vaccines:
Targeting and Exploiting the Bacterial
Quorum Sensing Pathway, Award No.
N00014-06-1-1176, and the National
Science Foundation International Materials
Institute Program for Combinatorial
Sciences and Materials Informatics
Collaboratory, Grant no. DMR-
0603644.
REFERENCES
1. R.D. King et al., Bioinformatics, 21 (2005), pp.
2017–2026.
2. E.C. Butcher et al., Nature Biotechnology, 22 (2004),
pp. 1253–1259.
3. A.K. Noor et al., Computers and Structures, 74
(2000), pp. 507–519.
4. B.D. Wirth et al., Nuclear Instruments and Methods
in Physics Research B, 180 (2001), pp. 23–31.
5. H. Kitano, Science, 295 (2002), pp. 1662–1664.
6. T. Ideker and D. Lauffenberger, Trends in
Biotechnology, 21 (2003), pp. 255–262.
7. C. Suh et al., “Informatics Methods for Combinatorial
Materials Science,” Combinatorial Materials Science,
ed. B. Narasimhan, S.K. Mallapragada, and M.D. Porter
(Hoboken, NJ: John Wiley, 2007), chapter 5.
8. C. Suh and K. Rajan, QSAR and Combinatorial
Sciences, 24 (2005), p. 114.
9. Zi-Kui Liu et al., JOM, 58 (11) (2006), pp. 42–50.
10. K. Rajan, Materials Today 8 (2005), pp. 38–45.
Krishna Rajan is with the Department of Materials
Science and Engineering, NSF Combinatorial
Sciences and Materials Informatics Collaboratory—
International Materials Institute (CoSMIC-IMI), Iowa
State University, Ames 50011. Dr. Rajan can be
reached at krajan@iastate.edu.
|